A research team, led by Professor Thorsten WOHLAND from both the Department of Biological Sciences and the Department of Chemistry, and Professor Adrian RÖLLIN, from the Department of Statistics and Data Science, both from NUS, used deep learning methods to reduce the amount of data required for a measurement (about 5 MB per measurement) while achieving comparable results to traditional methods. The technique uses two CNNs, called FCSNet and ImFCSNet developed by Dr Wai Hon TANG and Mr Shao Ren SIM, members of the research team. CNNs represent a type of deep learning algorithm suited for analysing visual data. They employ multiple layers of specialised filters that scan across the image for specific features, such as edges, textures and colours. By progressively extracting and combining these features together, they build up a better understanding of the image, allowing them to recognise patterns and objects within the visual data.
Both the FCSNet and ImFCSNet based methods are more precise than traditional FCS methods in terms of diffusion coefficients, but they have different trade-offs for data requirements and spatial resolution. By utilising less data, these techniques can potentially shorten the evaluation time by orders of magnitude, especially for large datasets or complex systems.
Their findings have been published in the Biophysical Journal.
Prof Wohland said, “These CNNs are trained on simulated data and can predict diffusion coefficients from much smaller datasets compared to traditional FCS methods. They are also model agnostic and can be used with any microscope setup.”
Dr Tang added, “CNNs revolutionise data analysis, offering accelerated and simplified evaluation processes. While it is important to validate their performance, CNNs have the potential to make powerful techniques available to the wider research community.”
The team hopes that their technique can open new possibilities to accelerate single-molecule research and make the technology accessible to a wider range of users. The CNNs do not require expert inputs and are poised to democratise FCS.
Read the full article here.
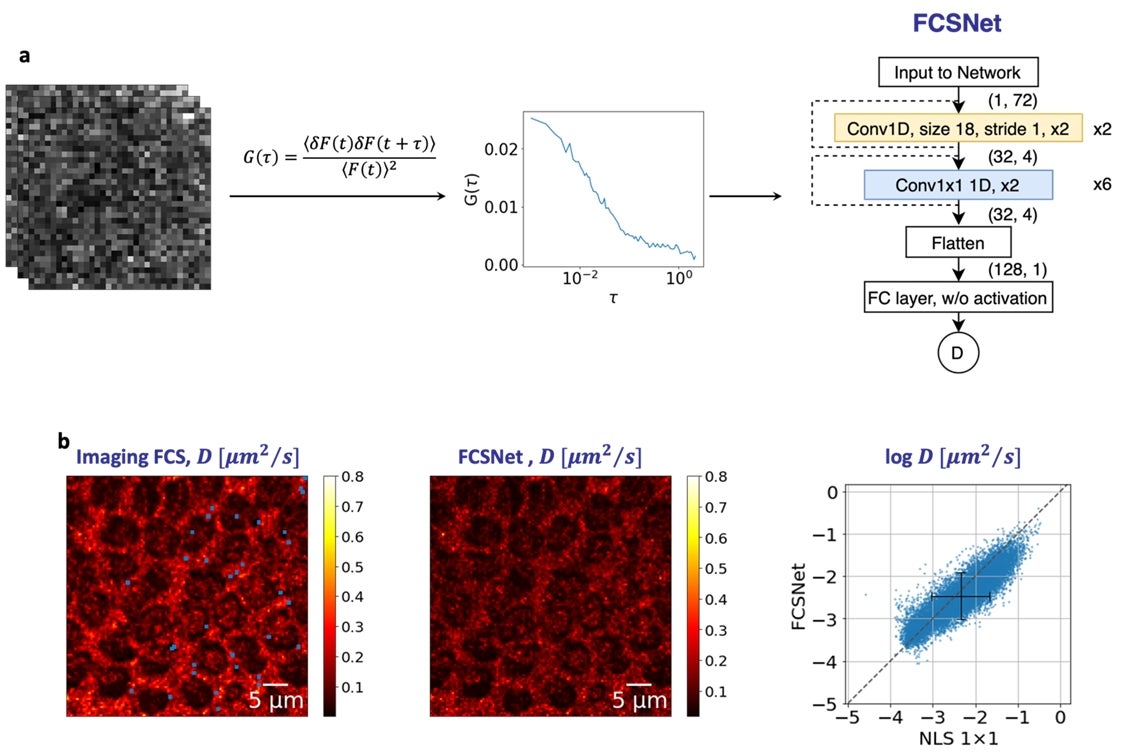
Figure: (a) FCSNet is a convolutional neural network (CNN), which takes in the autocorrelation function of a measurement as an input to predict the diffusion coefficient. (b) FCSNet allows evaluations in complex samples with a precision as good, or better than traditional Imaging FCS methods, as shown by the heatmaps. [Credit: Biophysical Journal]
