Research Fellow, Institute of Molecular and Cell Biology and Genome Institute of Singapore, A*STAR, 2015–2019; Ph.D. in Biophysics, Harvard University, 2009–2015; B.Sc. (honors) in Chemistry and Biology, Caltech, 2004–2008.
Contact Information:
Office: S1A-02-13
Tel: (65)-6516 4384
Email: zhaozw@nus.edu.sg
The research in my group intersects optical imaging, molecular biology and physical chemistry, with a specific focus on developing and applying advanced imaging-based methodologies to quantitatively probe the biophysics of chromatin dynamics in single human cells. In particular, we are interested in understanding how molecular processes that modulate genome organization, accessibility and expression are regulated in space and time. By integrating approaches from optical microscopy/spectroscopy (e.g. super-resolution imaging, single-molecule tracking and fluorescence correlation spectroscopy), omic technologies, correlative strategies and computational analysis, our work aims to illuminate the physico-chemical driving forces (such as biomolecular phase separation) that govern chromatin dynamics and cell nuclear architecture in vivo. Current efforts in the group are focused on chromatin remodeling and nuclear membrane–chromatin interactions, as well as the physiological implications of their misregulation in human health and diseases (particularly cancer and aging-associated disorders).
We are constantly looking for talented individuals from diverse backgrounds in life sciences, chemistry, biophysics, physics, bioengineering or related fields to join us.
Prospective postdoctoral fellows and Ph.D./M.Sc./undergraduate students please email Winston directly here.
We recently accomplished the first live-cell single-molecule quantification of the fully assembled human SWI/SNF chromatin remodeler complex, a critical family of multi-subunit protein complexes responsible for regulating genome access (via repositioning nucleosomes) in the cell and known to be implicated in >20% of all human cancers. We resolved temporally distinct modes of intranuclear diffusion and chromatin-binding, and uncovered a key role of the bromodomain (BD) in enhancing the chromatin-binding dynamics of the remodeler complex in a DNA-accessibility-dependent manner. Moreover, to furnish spatial contexts for such temporal dynamics, we devised a novel strategy (termed STAR) capable of super-resolved density mapping of single-molecule binding, and revealed spatially heterogenous, nanoscale remodeler binding “hotspots” across the cell nucleus where multiple binding events (especially consecutive longer-lived stable binding) preferentially cluster, hence leading to a model for the spatio-temporal organization of remodeler binding dynamics to selectively modulate genome access. Finally, through a systematic comparison of six common SWI/SNF remodeler mutants implicated in various cancers across tumor types, we revealed multi-modal changes unique to each mutant, thereby establishing the biophysical basis for aberrant remodeler–chromatin interactions that could potentially serve as a unique set of quantitative signatures for cancer-associated remodeler mutations.
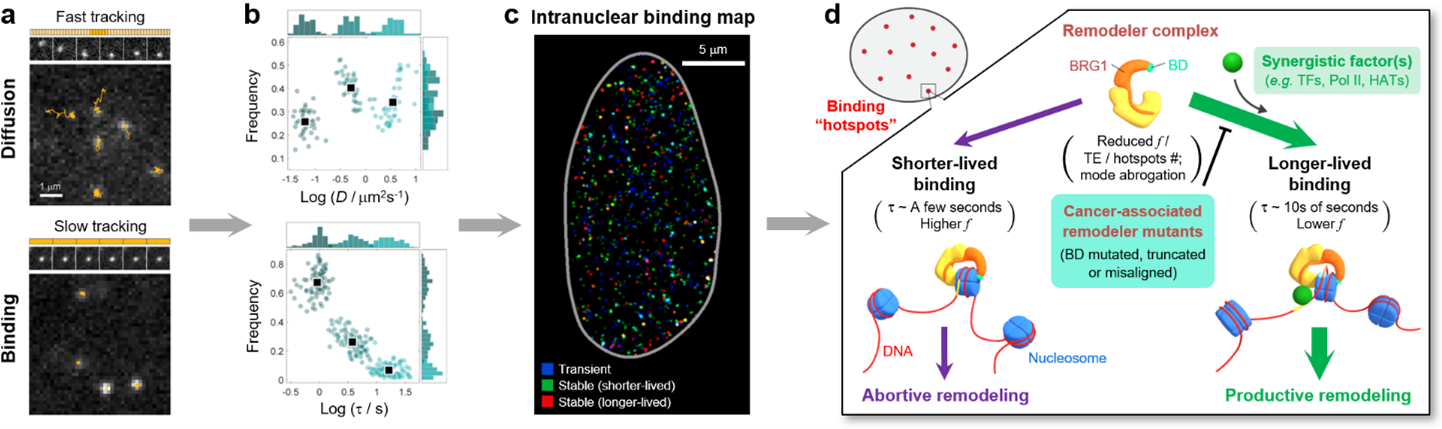
Figure: Live-cell single-molecule tracking of the fully assembled SWI/SNF chromatin remodeler complex (a) resolved temporally distinct modes of intranuclear diffusion and chromatin-binding (both transient and stable) (b). In addition, STAR mapping uncovered numerous nanoscale remodeler binding “hotspots” across the cell nucleus (c), pointing to an integrated structure-dynamics model (d), in which successive longer-lived binding of the remodeler complex within the “hotspots”, accompanied by the actions of synergistic factors, results in sustained productive remodeling activity at these sites. However, such longer-lived binding is drastically reduced (in terms of binding frequency (f), targeting efficiency (TE) or number of stable binding hotspots) or abrogated altogether for a variety of cancer-associated remodeler mutants, in which the BD is either mutated, truncated or misaligned in the remodeler–nucleosome complex. Ref: Engl, W. et al. Nat. Commun. 2024, 15, 7646.